Our projects
Yeast 2.0
The Yeast 2.0 project is a global partnership focused on utilising synthetic biology tools to build the world’s first synthetic eukaryotic
genome. The project has attracted key international leaders from institutions including New York University (USA), John Hopkins University (USA), the Joint Genome Institute (USA), BGI (China), Tianjin University (China), Tsinghua University (China), Imperial College London (UK), the University of Edinburgh (UK), and now Australia (hosted at Macquarie University) and will generate a synthetic genome for the well-studied yeast organism Saccharomyces cerevisiae (S. cerevisiae).

The synthetic yeast genome will be used to answer a wide variety of profound questions about fundamental properties of chromosomes, including:
- Genome organisation
- Function of RNA splicing
- The extent to which small RNAs play a role in yeast biology
- The distinction between prokaryotes and eukaryotes
- Questions relating to genome structure and evolution
The availability of a fully synthetic genome will allow direct testing of evolutionary questions not otherwise approachable. Yeast, and Saccharomyces cerevisiae in particular, are preeminent organisms for industrial fermentations, with a wide variety of practical uses including ethanol production from agricultural products and by-products.
 The Macquarie project team, in partnership with the Australian Wine Research Institute, have a good track record of productive collaborations in the disciplines required for project delivery and many years of experience in working with the target organisms. Together, we will synthesise chromosomes XIV and XVI as part of the global effort to generate a synthetic eukarytotic genome.
The Macquarie project team, in partnership with the Australian Wine Research Institute, have a good track record of productive collaborations in the disciplines required for project delivery and many years of experience in working with the target organisms. Together, we will synthesise chromosomes XIV and XVI as part of the global effort to generate a synthetic eukarytotic genome.
Macquarie University is excited to lead Australia into the next generation of scientific discovery!
Strategy
Building S.cerevisiae Chromosome XIV
Synthetic DNA is manufactured to incorporate several design features to be consistent throughout the genome of the synthetic yeast. Some of these features include the conversion of all stop codons to TAA and the insertion of loxPsym sites flanking all non-essential genes. In our laboratory we start with chunks of DNA up to 10 kb in length with “sticky” overhangs and co-transform our strain with a series of 4 chunks.
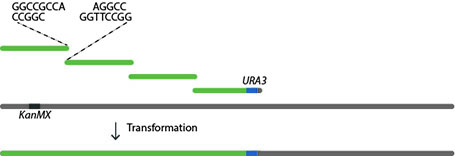
The end most chunk contains marker LEU2 or URA3, alternating between successive megachunks allowing selection of integration with each round.
Following phenotypic screening, the correct integration of synthetic DNA chunks is screened by real time PCR analysis. PCR Tag primers are designed to differentiate between wt and syn DNA. The entire length of the 750 kb chromosome will be made “synthetic” by this iterative process.
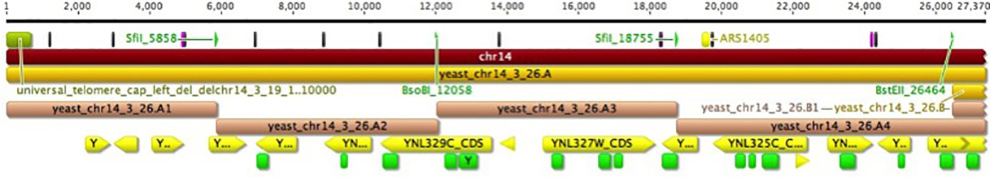
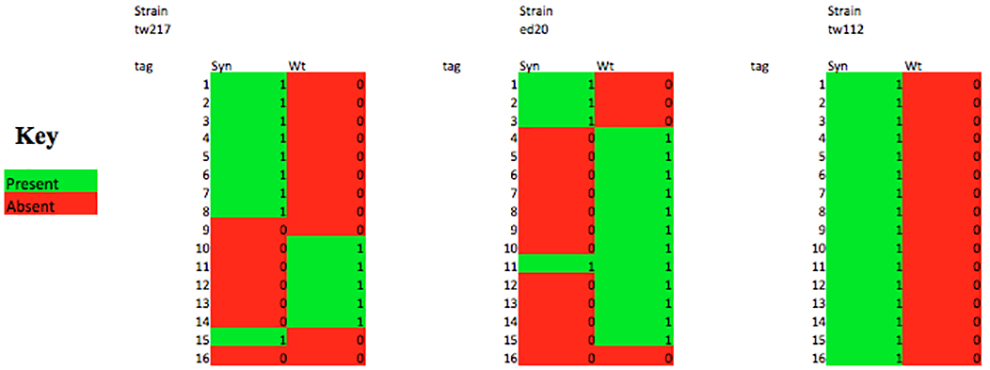
Figures by Tom Williams and Natalie Curach
Ethics
All research conducted within Macquarie University is subjected to scrutiny and approval by the Bioethics Committee. The yeast 2.0 project is part of a global consortium, which spans a number of local policy and practice boundaries in regard to ethics and governance. To ensure research is conducted is a uniform safe and ethical manner throughout the consortium, a specific Statement of Ethics has been developed. In it we declare research for the benefit of humankind, transparency and engagement, and consideration for the health and safety of people and the environment.